复现这篇文章基于随机森林方法的缺失值填充
导入各自包:
1
2
3
4
5
6
7
| import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# from sklearn.datasets import load_boston # 新版本无波士顿数据
from sklearn.impute import SimpleImputer # 填充缺失值的类
from sklearn.ensemble import RandomForestRegressor # 随机森林回归
from sklearn.model_selection import cross_val_score # 交叉验证
|
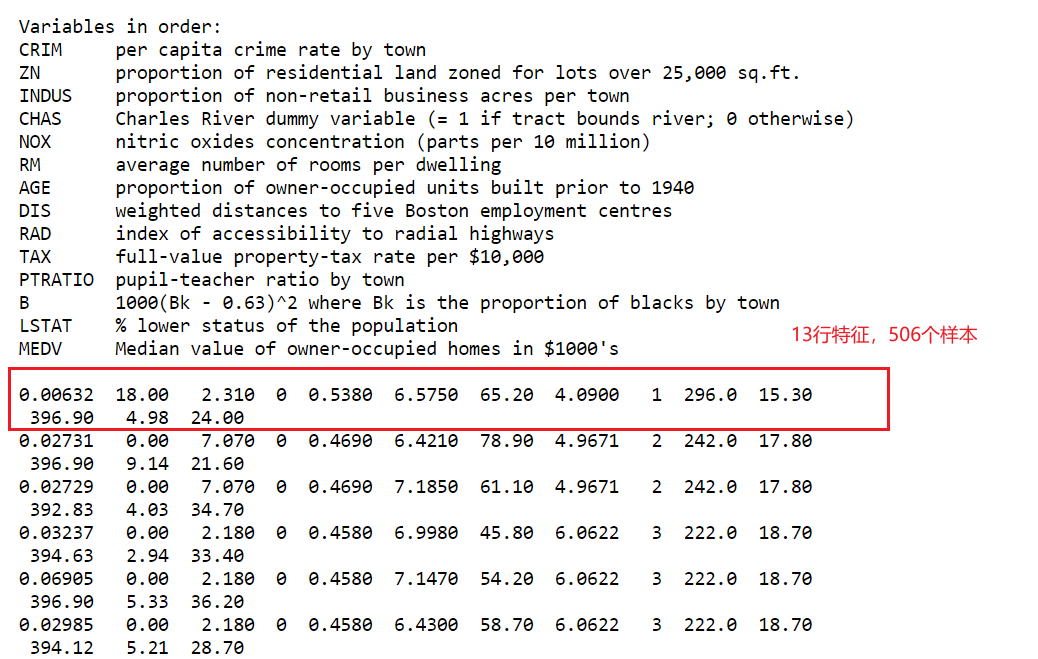
1 波士顿数据集获取
利用犯罪率、是否邻近查尔斯河、公路可达性等信息,来预测 20 世纪 70
年代波士顿地区房屋价格的中位数。这个数据集包含 506 个数据点和 13
个特征。波士顿房价数据集在scikit-learn1.2版本以后被移除了,通过其他网站获取,该数据已整理成excel,放置于:https://github.com/sdhlw/OA/tree/main/Boston_data
数据:前12行为特征即x,最后一行为y
1
2
3
4
5
| data_url = "http://lib.stat.cmu.edu/datasets/boston"
raw_df = pd.read_csv(data_url, sep="\s+", skiprows=22, header=None)
data = np.hstack([raw_df.values[::2, :], raw_df.values[1::2, :2]])
target = raw_df.values[1::2, 2] # 归一化和特征构造后都没有变化
print("Data shape: {}".format(data.shape))
|
2 设置缺失样本
1
2
3
4
5
| rng = np.random.RandomState(0)
missing_rate = 0.5
n_missing_samples = int(np.floor(n_samples * n_features * missing_rate))
print(n_missing_samples)
|
1
2
3
|
missing_features = rng.randint(0, n_features, n_missing_samples)
missing_samples = rng.randint(0, n_samples, n_missing_samples)
|
1
2
3
4
5
| X_missing = X_full.copy()
y_missing = y_full.copy()
X_missing[missing_samples,missing_features] = np.nan
X_missing = pd.D
|
3 数据填充
3.1 均值填充
1
2
3
| imp_mean = SimpleImputer(missing_values=np.nan, strategy="mean")
X_missing_mean = imp_mean.fit_transform(X_missing)
pd.DataFrame(X_missing_mean).isnull().sum()
|
3.2 0值填充
1
2
3
| imp_0 = SimpleImputer(missing_values=np.nan, strategy="constant", fill_value=0)
X_missing_0 = imp_0.fit_transform(X_missing)
pd.DataFrame(X_missing_0).isnull().sum()
|
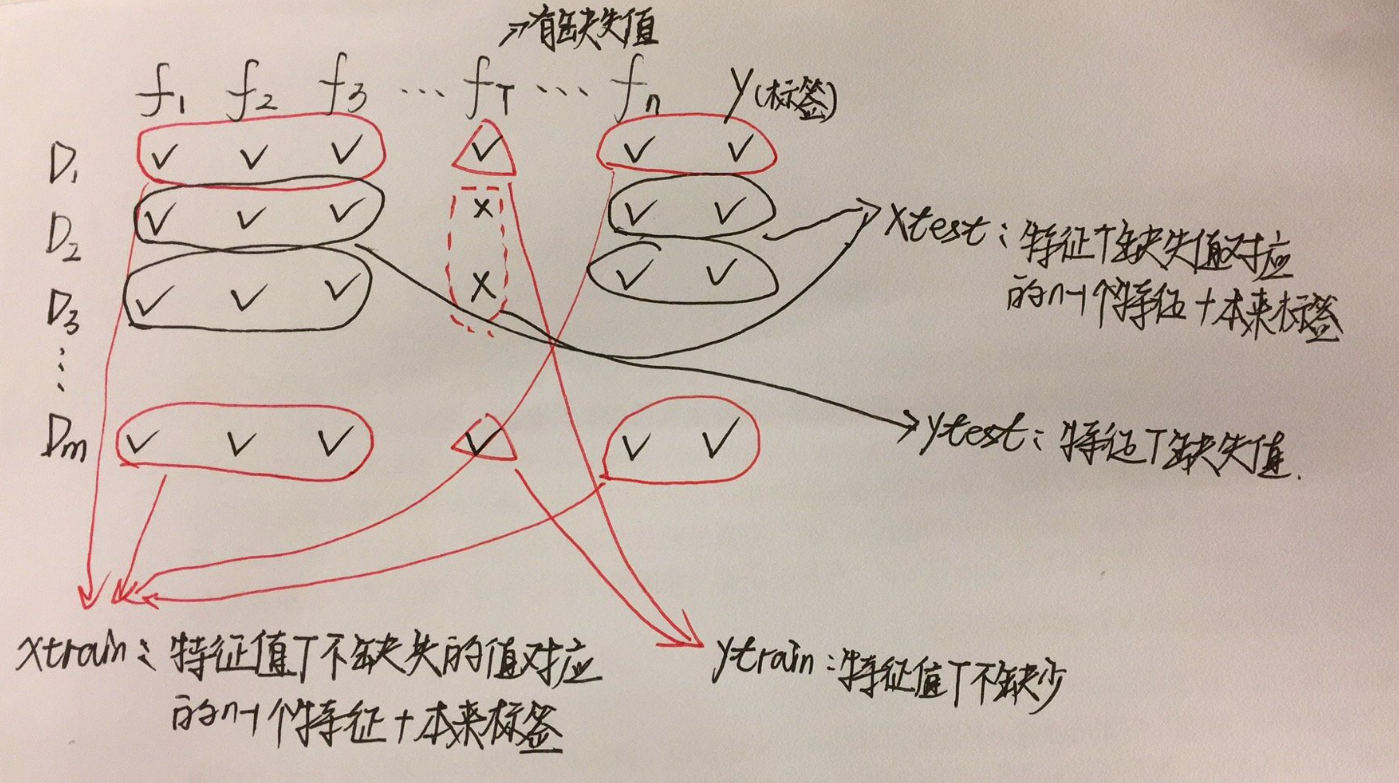
3.3 随机森林填充
假设一个具有n个特征的数据,特征T存在缺失值(大量缺失更适合),把T当做是标签,其他的n-1个特征和原来的数据看作是新的特征矩阵,具体数据解释为:
| Xtrain |
特征T不缺失的值对应的n-1个特征+原始标签 |
| ytrain |
特征T不缺失的值 |
| Xtest |
特征T缺失的值对应的n-1个特征+原始标签 |
| ytest |
特征T缺失值(未知) |
注意:
如果其他特征也存在缺失值,遍历所有的特征,从缺失值最少的开始。
- 缺失值越少,所需要的准确信息也越少
- 填补一个特征,先将其他特征值的缺失值用
0代替,这样每次循环一次,有缺失值的特征便会减少一个
假设数据有n个特征,m行数据:
由于是从最少的缺失值特征开始填充,那么需要找出存在缺失值的索引的顺序:argsort函数的使用

1
2
3
| X_missing_reg = X_missing.copy()
sortindex = np.argsort(pd.DataFrame(X_missing_reg).isnull().sum(axis=0)).values
|
填充过程:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
| for i in sortindex:
df = X_missing_reg
fillc = df.iloc[:, i]
df = pd.concat([df.iloc[:, df.columns != i], pd.DataFrame(y_full)], axis=1)
df_0 = SimpleImputer(missing_values=np.nan, strategy='constant', fill_value=0).fit_transform(df)
ytrain = fillc[fillc.notnull()]
ytest = fillc[fillc.isnull()]
Xtrain = df_0[ytrain.index, :]
Xtest = df_0[ytest.index, :]
rfc = RandomForestRegressor(n_estimators=100)
rfc = rfc.fit(Xtrain, ytrain)
y_predict = rfc.predict(Xtest)
X_missing_reg.loc[X_missing_reg.iloc[:, i].isnull(), i] = y_predict
|
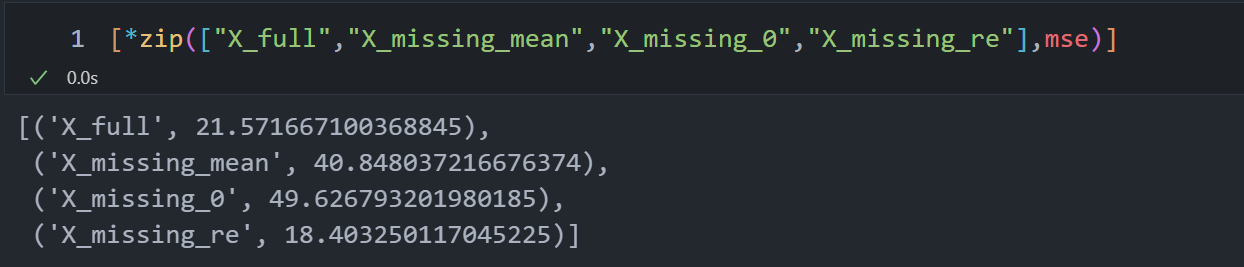
4 四种数据建模
- MSE:均方误差;回归树的衡量指标,越小越好。sklearn中使用的是负均方误差neg_mean_squared_error。均方误差本身是种误差loss,通过负数表示
- R2:回归树score返回的真实值是R的平方,不是MSE
1
2
3
4
5
6
7
| X = [X_full, X_missing_mean, X_missing_0, X_missing_reg]
mse = []
for x in X:
estimator = RandomForestRegressor(random_state=0, n_estimators=100)
scores = cross_val_score(estimator, x, y_full, scoring="neg_mean_squared_error", cv=5).mean()
mse.append(scores * -1)
|
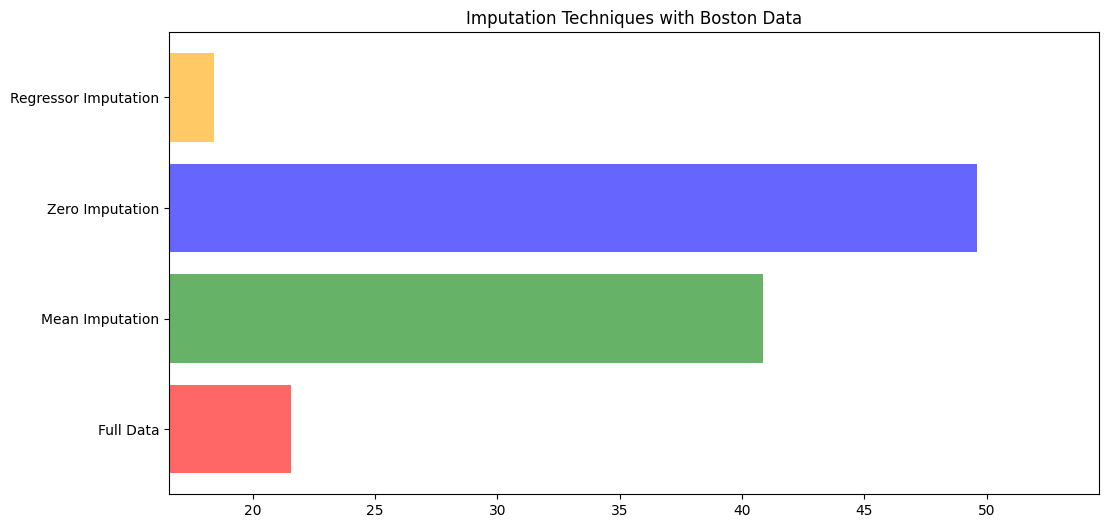
5 绘图
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
| x_labels = ["Full Data",
"Mean Imputation",
"Zero Imputation",
"Regressor Imputation"]
colors = ['r', 'g', 'b', 'orange']
plt.figure(figsize=(12,6))
ax = plt.subplot(111)
for i in np.arange(len(mse)):
ax.barh(i, mse[i], color=colors[i],alpha=0.6,align='center')
ax.set_title("Imputation Techniques with Boston Data")
ax.set_xlim(left=np.min(mse)*0.9,
right=np.max(mse)*1.1)
ax.set_yticks(np.arange(len(mse)))
ax.set_label("MSE")
ax.set_yticklabels(x_labels)
plt.show()
|
参考文章
无




